params <- list(
n_sims = 1000,
n_sites = 200,
n_sims_augmented = 200,
n_sites_augmented = 50,
n_pseudospecies_augmented = 50
)Overview
This document performs simulation-based calibration for the models
available in R package flocker. Here, our goal is to
validate flocker’s data formatting, decoding, and
likelihood implementations, and not brms’s construction of
the linear predictors.
The encoding of the data for a flocker model tends to be
more complex in the presence of missing observations, and so we include
missingness in the data simulation wherever possible (some visits
missing in all models, some time-steps missing in multiseason
models).
In all models, we include one unit covariate that affects detection and occupancy, colonization, extinction and/or autologistic terms as applicable, and one event covariate that affects detection only (for all models except the rep-constant).
Single-season
Rep-constant
# make the stancode
model_name <- paste0(tempdir(), "/sbc_rep_constant_model.stan")
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1,
params = list(
coefs = data.frame(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1)
)
),
seed = NULL,
rep_constant = TRUE,
ragged_rep = TRUE
)
flocker_data = make_flocker_data(fd$obs, fd$unit_covs, quiet = TRUE)
scode <- flocker_stancode(
f_occ = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1,
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", dpar = "occ"),
backend = "cmdstanr"
)
writeLines(scode, model_name)
rep_constant_generator <- function(N){
fd <- simulate_flocker_data(
n_pt = N, n_sp = 1,
params = list(
coefs = data.frame(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1)
)
),
seed = NULL,
rep_constant = TRUE,
ragged_rep = TRUE
)
flocker_data = make_flocker_data(fd$obs, fd$unit_covs, quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coefs$det_intercept,
`b[2]` = fd$params$coefs$det_slope_unit,
`b_occ[1]` = fd$params$coefs$occ_intercept,
`b_occ[2]` = fd$params$coefs$occ_slope_unit
),
generated = flocker_standata(
f_occ = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1,
flocker_data = flocker_data
)
)
}
rep_constant_gen <- SBC_generator_function(
rep_constant_generator,
N = params$n_sites
)
rep_constant_dataset <- suppressMessages(
generate_datasets(rep_constant_gen, params$n_sims)
)
rep_constant_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_rep_constant_model.stan")
)
)
rep_constant_results <- compute_SBC(rep_constant_dataset, rep_constant_backend)
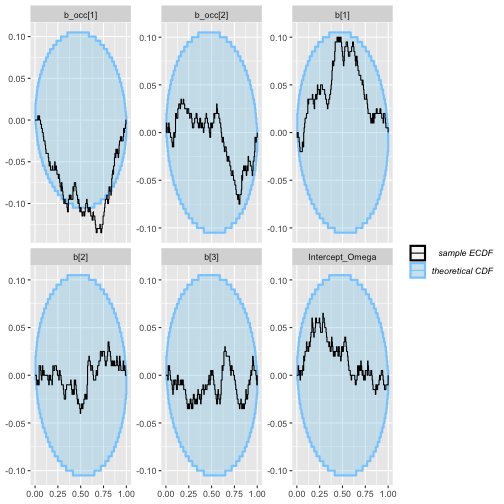
plot_ecdf(rep_constant_results)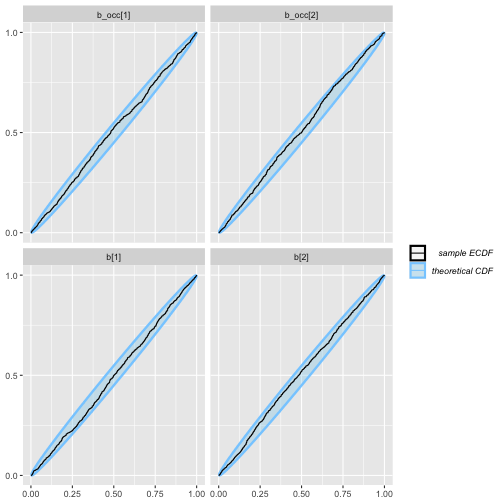
plot_rank_hist(rep_constant_results)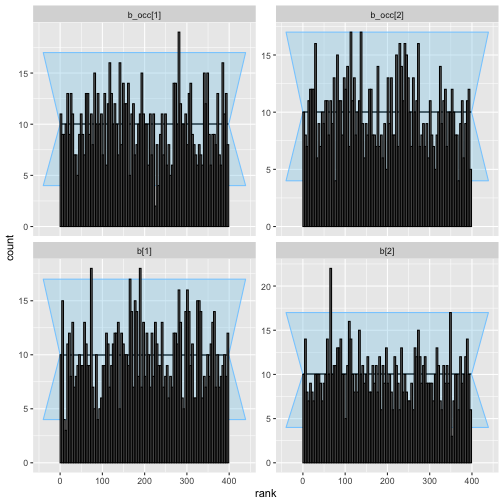
plot_ecdf_diff(rep_constant_results)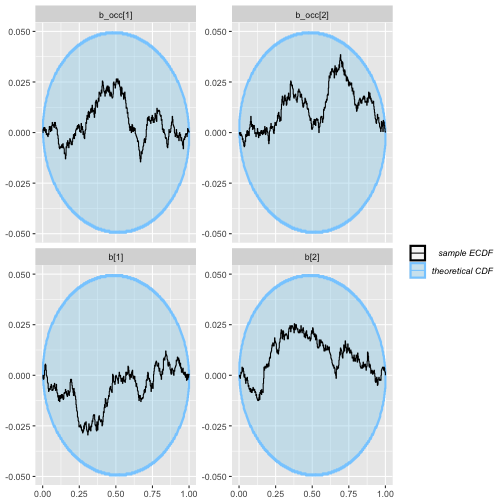
Rep-varying
# make the stancode
model_name <- paste0(tempdir(), "/sbc_rep_varying_model.stan")
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1,
params = list(
coefs = data.frame(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1)
)
),
seed = NULL,
rep_constant = FALSE,
ragged_rep = TRUE
)
flocker_data = make_flocker_data(fd$obs, fd$unit_covs, fd$event_covs, quiet = TRUE)
scode <- flocker_stancode(
f_occ = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", dpar = "occ"),
backend = "cmdstanr"
)
writeLines(scode, model_name)
rep_varying_generator <- function(N){
fd <- simulate_flocker_data(
n_pt = N, n_sp = 1,
params = list(
coefs = data.frame(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1)
)
),
seed = NULL,
rep_constant = FALSE,
ragged_rep = TRUE
)
flocker_data = make_flocker_data(fd$obs, fd$unit_covs, fd$event_covs, quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coefs$det_intercept,
`b[2]` = fd$params$coefs$det_slope_unit,
`b[3]` = fd$params$coefs$det_slope_visit,
`b_occ[1]` = fd$params$coefs$occ_intercept,
`b_occ[2]` = fd$params$coefs$occ_slope_unit
),
generated = flocker_standata(
f_occ = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data
)
)
}
rep_varying_gen <- SBC_generator_function(
rep_varying_generator,
N = params$n_sites
)
rep_varying_dataset <- suppressMessages(
generate_datasets(rep_varying_gen, params$n_sims)
)
rep_varying_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_rep_varying_model.stan")
)
)
rep_varying_results <- compute_SBC(rep_varying_dataset, rep_varying_backend)## - 3 (0%) fits had at least one Rhat > 1.01. Largest Rhat was 1.016.## Not all diagnostics are OK.
## You can learn more by inspecting $default_diagnostics, $backend_diagnostics
## and/or investigating $outputs/$messages/$warnings for detailed output from the backend.
plot_ecdf(rep_varying_results)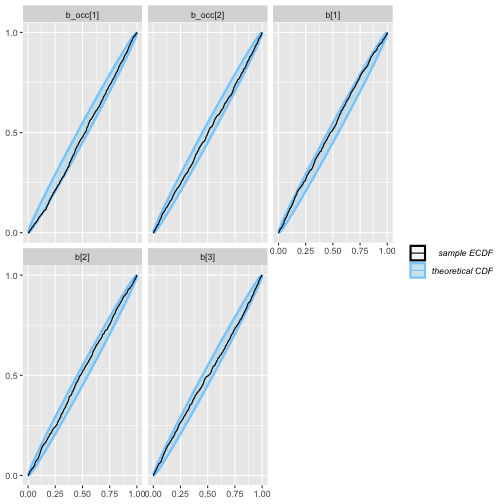
plot_rank_hist(rep_varying_results)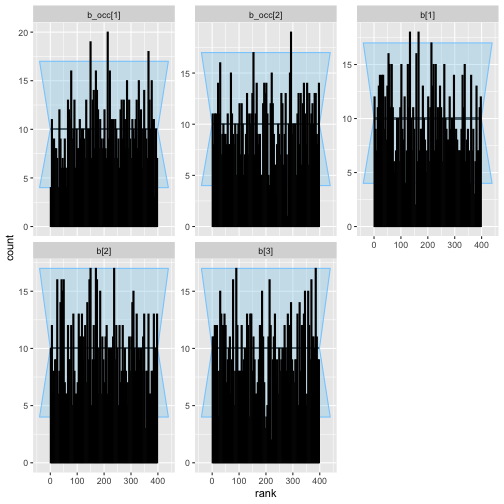
plot_ecdf_diff(rep_varying_results)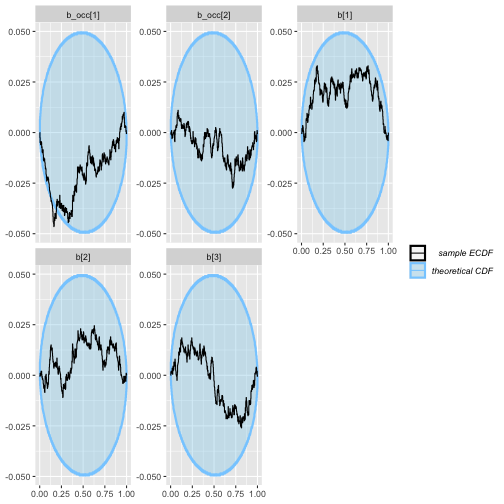
Multi-season
flocker fits multi-season models that parameterize the
dynamics using colonization/extinction or autologistic specifications,
and that parameterize the initial occupancy state using explicit and
equilibrium parameterizations, for a total of four classes of
multi-season model. We validate each class.
Colonization-extinction, explicit initial occupancy
# make the stancode
model_name <- paste0(tempdir(), "/sbc_colex_ex_model.stan")
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
coefs = data.frame(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1),
col_intercept = rnorm(1),
col_slope_unit = rnorm(1),
ex_intercept = rnorm(1),
ex_slope_unit = rnorm(1)
)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "colex",
multi_init = "explicit",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
scode <- flocker_stancode(
f_occ = ~ 0 + Intercept + uc1,
f_col = ~ 0 + Intercept + uc1,
f_ex = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", dpar = "occ") +
brms::set_prior("std_normal()", dpar = "colo") +
brms::set_prior("std_normal()", dpar = "ex"),
multiseason = "colex",
multi_init = "explicit",
backend = "cmdstanr"
)
writeLines(scode, model_name)
colex_ex_generator <- function(N){
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1),
colo_intercept = rnorm(1),
colo_slope_unit = rnorm(1),
ex_intercept = rnorm(1),
ex_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "colex",
multi_init = "explicit",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coefs$det_intercept,
`b[2]` = fd$params$coefs$det_slope_unit,
`b[3]` = fd$params$coefs$det_slope_visit,
`b_occ[1]` = fd$params$coefs$occ_intercept,
`b_occ[2]` = fd$params$coefs$occ_slope_unit,
`b_colo[1]` = fd$params$coefs$col_intercept,
`b_colo[2]` = fd$params$coefs$col_slope_unit,
`b_ex[1]` = fd$params$coefs$ex_intercept,
`b_ex[2]` = fd$params$coefs$ex_slope_unit
),
generated = flocker_standata(
f_occ = ~ 0 + Intercept + uc1,
f_col = ~ 0 + Intercept + uc1,
f_ex = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
multiseason = "colex",
multi_init = "explicit"
)
)
}
colex_ex_gen <- SBC_generator_function(
colex_ex_generator,
N = params$n_sites
)
colex_ex_dataset <- suppressMessages(
generate_datasets(colex_ex_gen, params$n_sims)
)
colex_ex_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_colex_ex_model.stan")
)
)
colex_ex_results <- compute_SBC(colex_ex_dataset, colex_ex_backend)
plot_ecdf(colex_ex_results)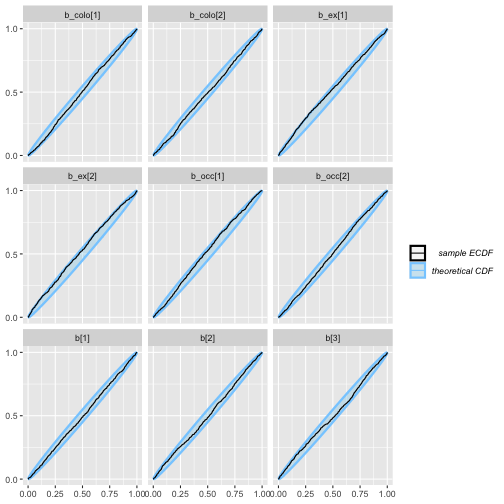
plot_rank_hist(colex_ex_results)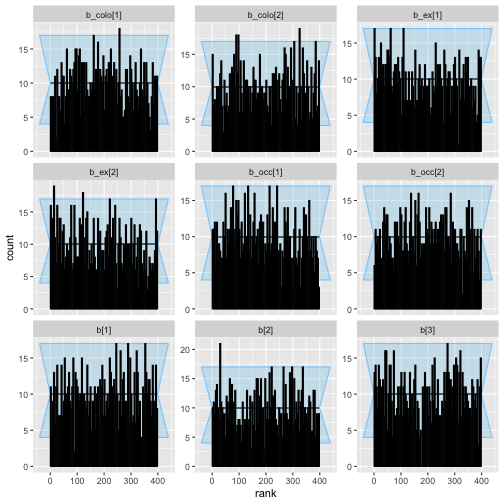
plot_ecdf_diff(colex_ex_results)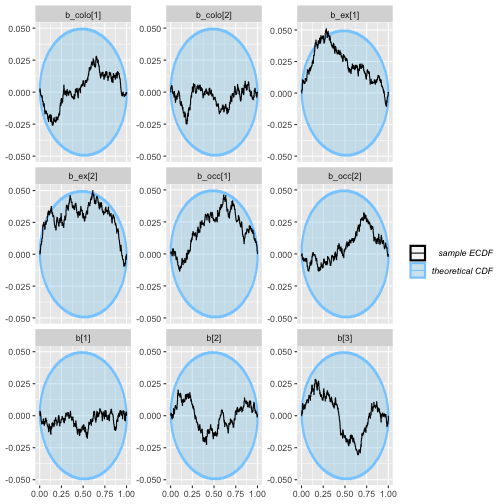
Colonization-extinction, equilibrium initial occupancy
# make the stancode
model_name <- paste0(tempdir(), "/sbc_colex_eq_model.stan")
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
colo_intercept = rnorm(1),
colo_slope_unit = rnorm(1),
ex_intercept = rnorm(1),
ex_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "colex",
multi_init = "equilibrium",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
scode <- flocker_stancode(
f_col = ~ 0 + Intercept + uc1,
f_ex = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", dpar = "colo") +
brms::set_prior("std_normal()", dpar = "ex"),
multiseason = "colex",
multi_init = "equilibrium",
backend = "cmdstanr"
)
writeLines(scode, model_name)
colex_eq_generator <- function(N){
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
col_intercept = rnorm(1),
col_slope_unit = rnorm(1),
ex_intercept = rnorm(1),
ex_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "colex",
multi_init = "equilibrium",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coefs$det_intercept,
`b[2]` = fd$params$coefs$det_slope_unit,
`b[3]` = fd$params$coefs$det_slope_visit,
`b_colo[1]` = fd$params$coefs$col_intercept,
`b_colo[2]` = fd$params$coefs$col_slope_unit,
`b_ex[1]` = fd$params$coefs$ex_intercept,
`b_ex[2]` = fd$params$coefs$ex_slope_unit
),
generated = flocker_standata(
f_col = ~ 0 + Intercept + uc1,
f_ex = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
multiseason = "colex",
multi_init = "equilibrium"
)
)
}
colex_eq_gen <- SBC_generator_function(
colex_eq_generator,
N = params$n_sites
)
colex_eq_dataset <- suppressMessages(
generate_datasets(colex_eq_gen, params$n_sims)
)
colex_eq_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_colex_eq_model.stan")
)
)
colex_eq_results <- compute_SBC(colex_eq_dataset, colex_eq_backend)## - 1 (0%) fits had at least one Rhat > 1.01. Largest Rhat was 1.012.## - 853 (85%) fits had some steps rejected. Maximum number of rejections was 5.## Not all diagnostics are OK.
## You can learn more by inspecting $default_diagnostics, $backend_diagnostics
## and/or investigating $outputs/$messages/$warnings for detailed output from the backend.
plot_ecdf(colex_eq_results)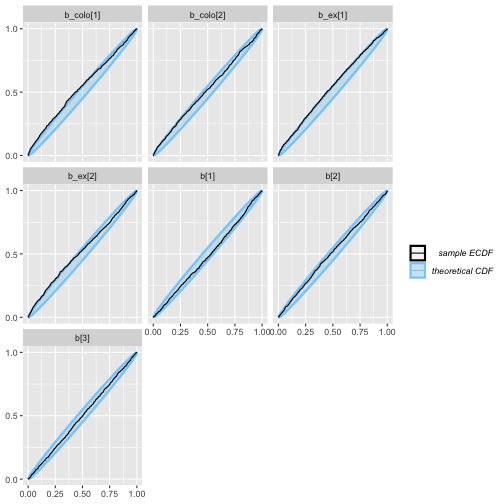
plot_rank_hist(colex_eq_results)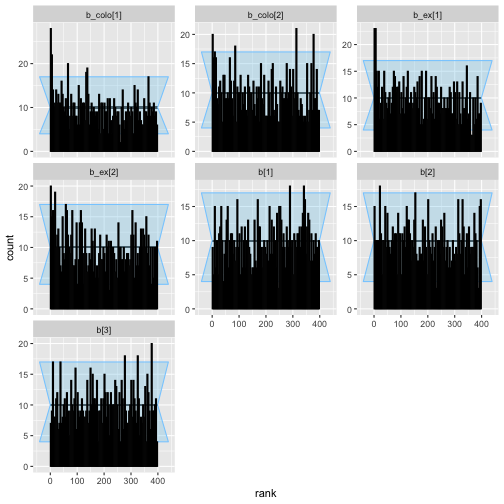
plot_ecdf_diff(colex_eq_results)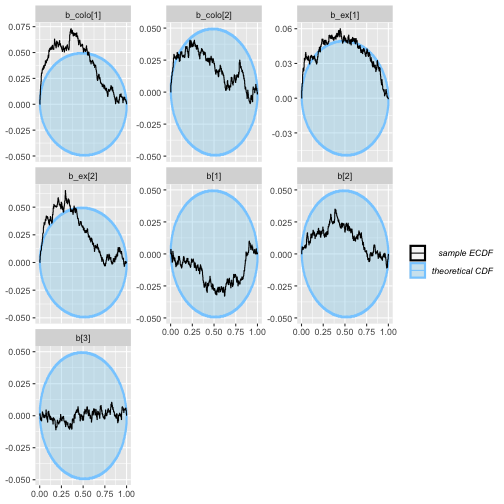
Autologistic, explicit initial occupancy
# make the stancode
model_name <- paste0(tempdir(), "/sbc_auto_ex_model.stan")
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1),
col_intercept = rnorm(1),
col_slope_unit = rnorm(1),
auto_intercept = rnorm(1),
auto_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "autologistic",
multi_init = "explicit",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
scode <- flocker_stancode(
f_occ = ~ 0 + Intercept + uc1,
f_col = ~ 0 + Intercept + uc1,
f_auto = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", dpar = "occ") +
brms::set_prior("std_normal()", dpar = "colo") +
brms::set_prior("std_normal()", dpar = "autologistic"),
multiseason = "autologistic",
multi_init = "explicit",
backend = "cmdstanr"
)
writeLines(scode, model_name)
auto_ex_generator <- function(N){
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
occ_intercept = rnorm(1),
occ_slope_unit = rnorm(1),
colo_intercept = rnorm(1),
colo_slope_unit = rnorm(1),
auto_intercept = rnorm(1),
auto_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "autologistic",
multi_init = "explicit",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coefs$det_intercept,
`b[2]` = fd$params$coefs$det_slope_unit,
`b[3]` = fd$params$coefs$det_slope_visit,
`b_occ[1]` = fd$params$coefs$occ_intercept,
`b_occ[2]` = fd$params$coefs$occ_slope_unit,
`b_colo[1]` = fd$params$coefs$col_intercept,
`b_colo[2]` = fd$params$coefs$col_slope_unit,
`b_autologistic[1]` = fd$params$coefs$auto_intercept,
`b_autologistic[2]` = fd$params$coefs$auto_slope_unit
),
generated = flocker_standata(
f_occ = ~ 0 + Intercept + uc1,
f_col = ~ 0 + Intercept + uc1,
f_auto = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
multiseason = "autologistic",
multi_init = "explicit"
)
)
}
auto_ex_gen <- SBC_generator_function(
auto_ex_generator,
N = params$n_sites
)
auto_ex_dataset <- suppressMessages(
generate_datasets(auto_ex_gen, params$n_sims)
)
auto_ex_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_auto_ex_model.stan")
)
)
auto_ex_results <- compute_SBC(auto_ex_dataset, auto_ex_backend)
plot_ecdf(auto_ex_results)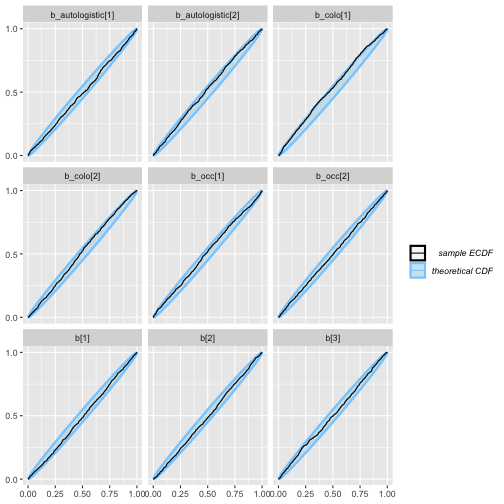
plot_rank_hist(auto_ex_results)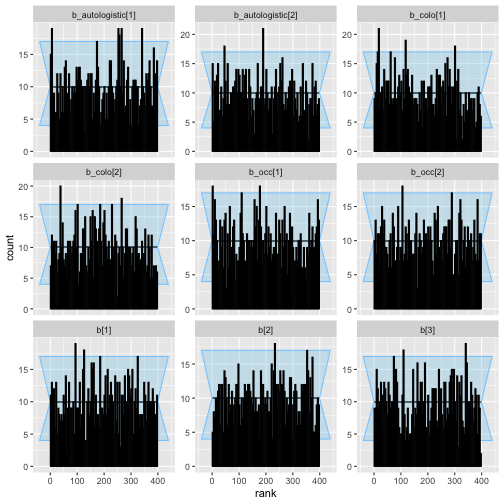
plot_ecdf_diff(auto_ex_results)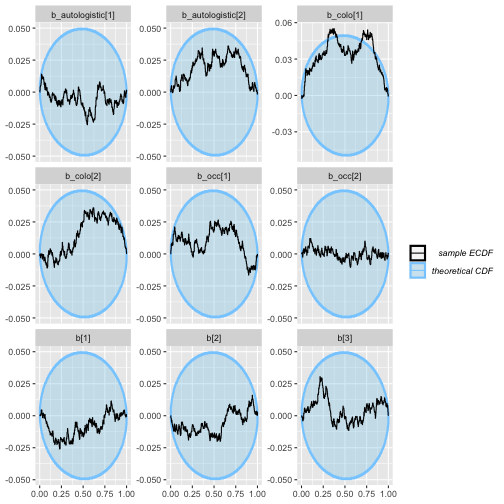
Autologistic, equilibrium initial occupancy
# make the stancode
model_name <- paste0(tempdir(), "/sbc_auto_eq_model.stan")
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
auto_intercept = rnorm(1),
auto_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "autologistic",
multi_init = "equilibrium",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
scode <- flocker_stancode(
f_col = ~ 0 + Intercept + uc1,
f_auto = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", dpar = "colo") +
brms::set_prior("std_normal()", dpar = "autologistic"),
multiseason = "autologistic",
multi_init = "equilibrium",
backend = "cmdstanr"
)
writeLines(scode, model_name)
auto_eq_generator <- function(N){
fd <- simulate_flocker_data(
n_pt = params$n_sites, n_sp = 1, n_season = 4,
params = list(
det_intercept = rnorm(1),
det_slope_unit = rnorm(1),
det_slope_visit = rnorm(1),
col_intercept = rnorm(1),
col_slope_unit = rnorm(1),
auto_intercept = rnorm(1),
auto_slope_unit = rnorm(1)
),
seed = NULL,
rep_constant = FALSE,
multiseason = "autologistic",
multi_init = "equilibrium",
ragged_rep = TRUE
)
flocker_data = make_flocker_data(
fd$obs, fd$unit_covs, fd$event_covs,
type = "multi", quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coefs$det_intercept,
`b[2]` = fd$params$coefs$det_slope_unit,
`b[3]` = fd$params$coefs$det_slope_visit,
`b_colo[1]` = fd$params$coefs$col_intercept,
`b_colo[2]` = fd$params$coefs$col_slope_unit,
`b_autologistic[1]` = fd$params$coefs$auto_intercept,
`b_autologistic[2]` = fd$params$coefs$auto_slope_unit
),
generated = flocker_standata(
f_col = ~ 0 + Intercept + uc1,
f_auto = ~ 0 + Intercept + uc1,
f_det = ~ 0 + Intercept + uc1 + ec1,
flocker_data = flocker_data,
multiseason = "autologistic",
multi_init = "equilibrium"
)
)
}
auto_eq_gen <- SBC_generator_function(
auto_eq_generator,
N = params$n_sites
)
auto_eq_dataset <- suppressMessages(
generate_datasets(auto_eq_gen, params$n_sims)
)
auto_eq_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_auto_eq_model.stan")
)
)
auto_eq_results <- compute_SBC(auto_eq_dataset, auto_eq_backend)## - 302 (30%) fits had some steps rejected. Maximum number of rejections was 3.## Not all diagnostics are OK.
## You can learn more by inspecting $default_diagnostics, $backend_diagnostics
## and/or investigating $outputs/$messages/$warnings for detailed output from the backend.
plot_ecdf(auto_eq_results)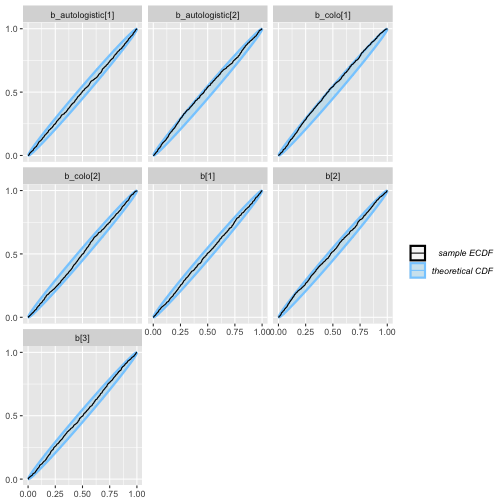
plot_rank_hist(auto_eq_results)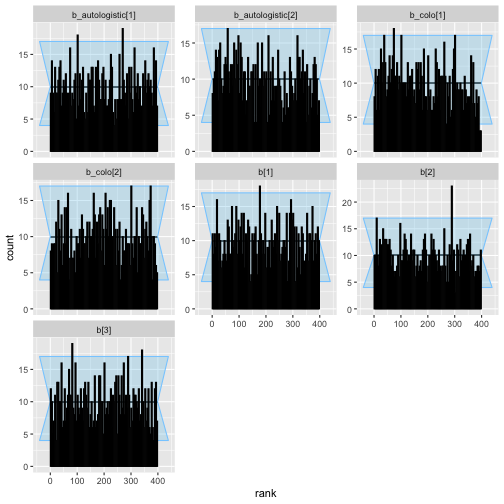
plot_ecdf_diff(auto_eq_results)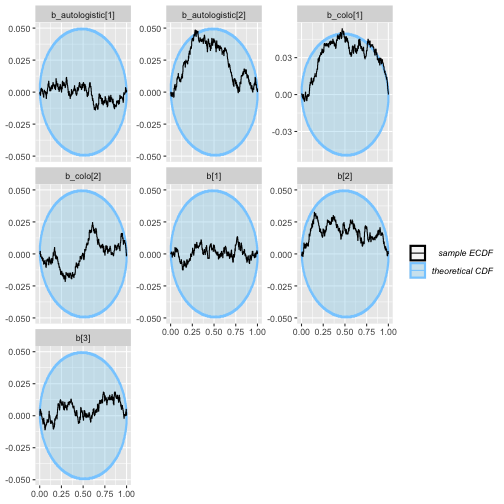
Data-augmented
# make the stancode
model_name <- paste0(tempdir(), "/sbc_augmented_model.stan")
omega <- boot::inv.logit(rnorm(1, 0, .1))
available <- rbinom(1, params$n_pseudospecies_augmented, omega)
unavailable <- params$n_pseudospecies_augmented - available
coef_means <- rnorm(5) # normal prior on random effect means
sigma <- abs(rnorm(5)) # half-normal prior on all random effect sds
Sigma <- diag(5) * sigma
fd <- simulate_flocker_data(
n_pt = params$n_sites_augmented, n_sp = available,
params = list(
coef_means = coef_means,
Sigma = Sigma,
coefs = data.frame(
det_intercept = rnorm(available, coef_means[1], sigma[1]),
det_slope_unit = rnorm(available, coef_means[2], sigma[2]),
det_slope_visit = rnorm(available, coef_means[3], sigma[3]),
occ_intercept = rnorm(available, coef_means[4], sigma[4]),
occ_slope_unit = rnorm(available, coef_means[5], sigma[5])
)
),
seed = NULL,
rep_constant = FALSE,
ragged_rep = TRUE
)
obs_aug <- fd$obs[seq_len(params$n_sites_augmented), ]
for(i in 2:available){
obs_aug <- abind::abind(
obs_aug,
fd$obs[((i - 1) * params$n_sites_augmented) + seq_len(params$n_sites_augmented), ],
along = 3
)
}
event_covs_aug <- list(ec1 = fd$event_covs$ec1[seq_len(params$n_sites_augmented), ])
unit_covs_aug <- data.frame(uc1 = fd$unit_covs[seq_len(params$n_sites_augmented), "uc1"])
flocker_data = make_flocker_data(
obs_aug, unit_covs_aug, event_covs_aug,
type = "augmented", n_aug = unavailable,
quiet = TRUE)
scode <- flocker_stancode(
f_occ = ~ 0 + Intercept + uc1 + (1 + uc1 || ff_species),
f_det = ~ 0 + Intercept + uc1 + ec1 + (1 + uc1 + ec1 || ff_species),
flocker_data = flocker_data,
prior =
brms::set_prior("std_normal()") +
brms::set_prior("std_normal()", class = "sd") +
brms::set_prior("std_normal()", dpar = "occ") +
brms::set_prior("std_normal()", class = "sd", dpar = "occ") +
brms::set_prior("normal(0, 0.1)", class = "Intercept", dpar = "Omega"),
backend = "cmdstanr",
augmented = TRUE
)
writeLines(scode, model_name)
aug_generator <- function(N){
omega <- boot::inv.logit(rnorm(1, 0, .1))
available <- rbinom(1, params$n_pseudospecies_augmented, omega)
unavailable <- params$n_pseudospecies_augmented - available
coef_means <- rnorm(5) # normal prior on random effect means
sigma <- abs(rnorm(5)) # half-normal prior on all random effect sds
Sigma <- diag(5) * sigma
fd <- simulate_flocker_data(
n_pt = params$n_sites_augmented, n_sp = available,
params = list(
coef_means = coef_means,
Sigma = Sigma,
coefs = data.frame(
det_intercept = rnorm(available, coef_means[1], sigma[1]),
det_slope_unit = rnorm(available, coef_means[2], sigma[2]),
det_slope_visit = rnorm(available, coef_means[3], sigma[3]),
occ_intercept = rnorm(available, coef_means[4], sigma[4]),
occ_slope_unit = rnorm(available, coef_means[5], sigma[5])
)
),
seed = NULL,
rep_constant = FALSE,
ragged_rep = TRUE
)
obs_aug <- fd$obs[seq_len(params$n_sites_augmented), ]
for(i in 2:available){
obs_aug <- abind::abind(
obs_aug,
fd$obs[((i - 1) * params$n_sites_augmented) + seq_len(params$n_sites_augmented), ],
along = 3
)
}
event_covs_aug <- list(ec1 = fd$event_covs$ec1[seq_len(params$n_sites_augmented), ])
unit_covs_aug <- data.frame(uc1 = fd$unit_covs[seq_len(params$n_sites_augmented), "uc1"])
flocker_data = make_flocker_data(
obs_aug, unit_covs_aug, event_covs_aug,
type = "augmented", n_aug = unavailable,
quiet = TRUE)
# format for return
list(
variables = list(
`b[1]` = fd$params$coef_means[1],
`b[2]` = fd$params$coef_means[2],
`b[3]` = fd$params$coef_means[3],
`b_occ[1]` = fd$params$coef_means[4],
`b_occ[2]` = fd$params$coef_means[5],
`Intercept_Omega` = boot::logit(omega)
),
generated = flocker_standata(
f_occ = ~ 0 + Intercept + uc1 + (1 + uc1 || ff_species),
f_det = ~ 0 + Intercept + uc1 + ec1 + (1 + uc1 + ec1 || ff_species),
flocker_data = flocker_data,
augmented = TRUE
)
)
}
aug_gen <- SBC_generator_function(
aug_generator,
N = params$n_sites_augmented
)
aug_dataset <- suppressMessages(
generate_datasets(aug_gen, params$n_sims_augmented)
)
aug_backend <-
SBC_backend_cmdstan_sample(
cmdstanr::cmdstan_model(
paste0(tempdir(), "/sbc_augmented_model.stan")
)
)
aug_results <- compute_SBC(aug_dataset, aug_backend)## - 4 (2%) fits had at least one Rhat > 1.01. Largest Rhat was 1.022.## - 2 (1%) fits had divergent transitions. Maximum number of divergences was 2.## - 7 (4%) fits had some steps rejected. Maximum number of rejections was 1.## Not all diagnostics are OK.
## You can learn more by inspecting $default_diagnostics, $backend_diagnostics
## and/or investigating $outputs/$messages/$warnings for detailed output from the backend.
plot_ecdf(aug_results)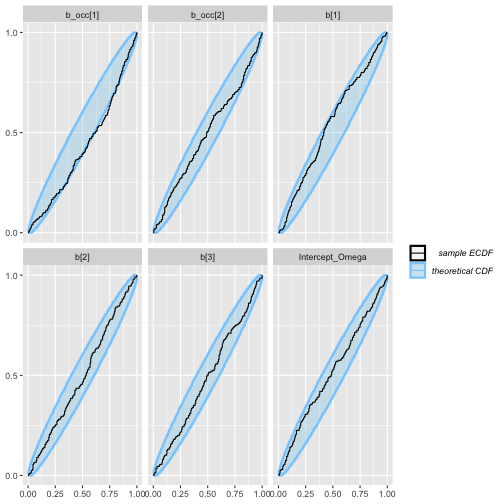
plot_rank_hist(aug_results)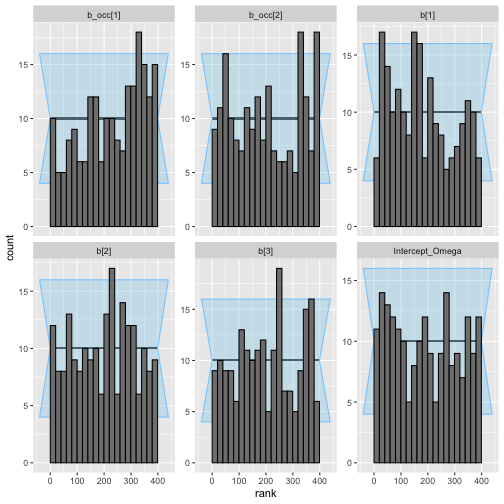
plot_ecdf_diff(aug_results)